The current HIV viral panels are outdated, poorly characterized and do not represent the diversity of viral strains observed worldwide. Furthermore, as clades have spread to new geographic regions, mutated and recombined, new circulating recombinant forms (CRFs) of the virus have emerged. This has necessitated the development of a well-characterized, global HIV isolate repository. To this end, one of the goals of EQAPOL is to establish and characterize unique clade-specific HIV virus panels.
The goal of the program is to establish a set of fully characterized viruses from early acute HIV infections that are consistent with the degree of viral evolution present globally for the following applications:
- Developing new assays
- Validating assay platforms
- Assisting regulators to evaluate test kits
- Monitoring HIV drug resistance
- Informing vaccine development
Through partnerships with the Blood Systems Research Institute and other collaborating sites, samples are gathered from newly-identified HIV-infected individuals with rare, unique or difficult to identify isolates, which are then be propagated to high titer in culture. To increase geographic diversity of the panel, previously cultured HIV has been gathered for further propagation by EQAPOL. All virus sources are cultured to high titer/high volume and then stored as neat culture supernatant, as well as spiked into defibrinated plasma at three different concentrations. Viruses are evaluated, characterized, and fully sequenced before adding them to a central repository where they will be available to the research community. For the EQAPOL Viral Diversity Program, 50 new viruses are cultured to high titer/high volume each year.
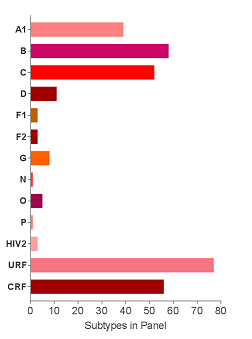
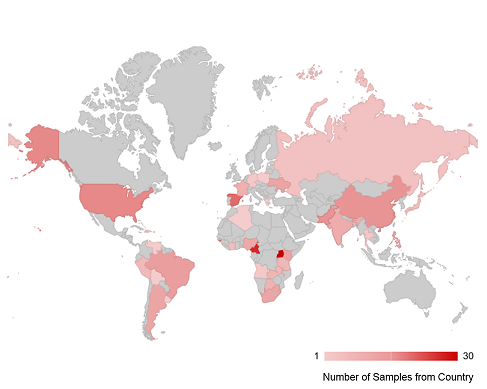
The current EQAPOL Viral Diversity panel includes over 322 viral isolates representing 6 subtypes (A, B, C, D, F, G), 2 sub-subtypes (F1, F2), 10 circulating recombinant forms (CRFs) (01, 02, 04, 11, 14, 22, 24, 47, 59, 70), 75 unique recombinant forms (URFs), 3 HIV-2, 1 group P, and 5 group O viruses from 37 countries as shown in the map below. More information about our other specialized panels is available here.
Viral Diversity samples are available to NIAID-approved laboratories. Products created for the EQAPOL Viral Diversity Program are available to order. These products can be requested via a secure, web-based application. Laboratories wishing to participate in the program should contact us.
As part of the EQAPOL Viral Diversity Panel, we are collaborating with groups to create specialized panels for a variety of uses. Panels are available for NIAID-approved sites. Below is a description of three specialized panels created using EQAPOL Viral Diversity Materials.
Genetic Diversity Panel: This panel consists of 50 EQAPOL viruses representing genetic and geographic diversity including samples with Drug Resistance Mutations. Panel viruses are spiked into defibrinated plasma and are available as a blinded panel. The panel is being produced in collaboration with Blood Systems Research Institute. The panel is being utilized by the Recipient Epidemiology and Donor Evaluation Study-III (REDS-III) project for validating genotyping methods.
Pseudotyped Virus Panel: The goal of this project is to develop a panel of standardized Env-pseudoviruses. The panel will be used primarily by Julie Overbaugh and other investigators in the HIV Superinfection Working Group. Material for generating the pseudoviruses was provided by the HIV Superinfection Working Group and Dr. David Montefiori from Duke University. The panel is available in 0.5 mL aliquots to interested sites.
Drug Resistance Virus Panel: This panel consists of 29 EQAPOL viruses with drug resistance mutations. These viruses are derived from different geographic regions and represent diverse virus strains. The near full-length genomes of the panel viruses are sequenced. The percentages of minor and major drug resistance mutations in each viral isolate are determined by next-generation sequencing.
- Subtypes: CRF01, CRF02, CRF14, CRF47, A1, B, C, D, F1, F2, G, O, URF_BC, URF_ADG, URF_A1B
- Countries of Origin: China, Cameroon, Brazil, Spain, USA, Uganda, Guinea
We are grateful for the following collaborators who have contributed source material to the EQAPOL Viral Diversity Program:
- Blood Systems Research Institute: Michael Busch, MD, PhD, Mars Stone, PhD and Sheila Keating, PhD
- Aga Khan University and Bridge Consultants Foundation: Rumina Hasan and Sharaf Ali
- Cureline, Inc.: Harminder Singh
- Harvard University: Vladimir Novitsky
- Nagoya Medical Center: Wataru Sugiura
- HIV Vaccine Trial Network (HVTN)
- American Red Cross: Susan Stramer, PhD
- South African National Blood Service: Marion Vermeulen
- Institut National de la Transfusion Sanguine: Syria Laperche, MD
- US Military HIV Research Program (MHRP), Henry M. Jackson Foundation: Mark Manak, PhD
- German Red Cross Institute Baden-Württemberg – Hessen: Michael Schmidt
- University of Sao Paulo: Ester Cerdeira Sabino, MD, PhD
- Japanese Red Cross: Kenji Tadokoro, MD
- Food and Drug Administration, Center for Biologics Evaluation and Research:Indira K. Hewlett, PhD and Viswanath Ragupathy, PhD
- Instituto de Salud Carlos III: Lucía Pérez-Álvarez, Elena Delgado, Michael Thomson, Aurora Fernández-García and Teresa Cuevas
- First Affiliated Hospital of China Medical University: Hong Shang, Wenqing Geng, Hualu Cui, and Hong Sun
- International AIDS Vaccine Initiative (IAVI): Pat Fast, MD, PhD, Jill Gilmour, BSc, PhD, Matt Price, PhD and Protocol C investigators
- University of Pennsylvania: George Shaw, MD, PhD, Beatrice Hahn, MD
- The Fraunhofer Institute for Biomedical Engineering IBMT: Hagen von Briesen, PhD
- University of Alabama at Birmingham: Joshua Baalwa
- National AIDS Research Institute, India: Ramesh Paranjape, PhD and Smita Kulkarni, PhD
- Duke Human Vaccine Institute: David Montefiori, PhD
- Center for HIV-AIDs Vaccine Immunology: Barton Haynes, MD
The EQAPOL Virus Diversity panel is composed of high/titer cultured viruses representing the geographic and genetic diversity of HIV. HIV sources are cultured to high titer/high volume and then stored as neat culture supernatant, as well as spiked into defibrinated plasma at two different concentrations. Viruses are evaluated, characterized, and fully sequenced before adding them to a central repository where they will be available to the research community. For the EQAPOL Viral Diversity Program, 25 new viruses are cultured to high titer/high volume each year.
You can find out the current inventory available from EQAPOL and request samples. Samples can be shipped to NIAID-approved laboratories. There is no cost of the samples, only the cost of shipping. Email your request.